Division of Gene Technology and Biomedicine
The Division of Gene Technology and Biomedicine continues the legacy of the TalTech Department of Gene Technology (founded in 1997), which in turn emerged on the basis of the Molecular Genetics Laboratory at the Institute of Chemical Physics and Biophysics (founded in 1980 and located in Tartu, from 1984 in Tallinn) and a group of researchers from the Institute of Experimental Biology at the Estonian University of Life Sciences (then named Estonian University of Agriculture). Our core competences are centered around mammalian biochemistry, molecular biology, and microbiology, with a strong emphasis on preclinical biomedical research and technological development. Currently the division hosts research groups working on lipid biochemistry, bioinformatics, microbiology, DNA replication, immunology, metalloproteomics, molecular neuroscience, reproductive biology, microfluidics and green analytical technologies for clinical and chemical forensic research. Our researchers teach courses at BSc, MSc and PhD levels. The division provides core facility and research services, including microscopy and animal facility infrastructure.
Pamphlet of the Division of Gene Technology and Biomedicine

Biochemistry of Lipids and Lipoproteins
The main topic of our research group is related to the identification of fundamental aspects of lipid and lipoprotein metabolism. Lipid and lipoprotein metabolism is associated with several health conditions, such as hypertriglyceridemia, cardiovascular diseases, pancreatitis, obesity and diabetes. Our main focus is studying the regulatory mechanisms of key enzymes (lipases) involved in lipid and lipoprotein metabolism. We have competence and experience in the study of the structure and properties of proteins, in enzymology and in the analysis of biomolecular interactions, as well as lipid analysis. In our research, we mainly apply calorimetry, chromatography, mass spectrometry, surface plasmon resonance and fluorescence-based technologies. Our work has led to industrial collaborations with companies such as Opocrin SPA (Italy) and Lipigon Pharmaceuticals (Sweden).
MAIN RESEARCH TOPICS INCLUDE:
- Studying the regulatory mechanisms of lipoprotein lipase activity
- Determining lipoprotein lipase activity in different human plasma environments
- Further development in pancreatic lipase and gastric lipase activity assays
- Development of an anti-hypertriglyceridemic drug formulation based on recombinant human lipoprotein lipase
MEMBERS OF THE RESEARCH GROUP:
Principal investigator: Aivar Lõokene
Group members: Robert Risti, Ly Villo, Ivar Järving
PhD students: Naatan Seeba
MSc students: Liise Hämarmets; Elizabet Solovjova
Post Docs: Mart Reimund

Patents: Method for calorimetric determination of the lipoprotein lipase activity in human plasma environment
CONTACT INFORMATION:
Aivar Lõokene, lead research scientist, head of the lipid and lipoprotein metabolism lab
E-mail: aivar.lookene@taltech.ee
Address: Building of Science, Akadeemia road 15, room 320
CV: https://www.etis.ee/CV/Aivar_L%C3%B5okene/eng/
- A negatively charged cluster in the disordered acidic domain of GPIHBP1 provides selectivity in the interaction with lipoprotein lipase: https://doi.org/10.1038/s41598-024-70468-6
- Combined action of albumin and heparin regulates lipoprotein lipase oligomerization, stability, and ligand interactions: https://doi.org/10.1371/journal.pone.0283358
- Lipoprotein Lipase Activity Does Not Differ in the Serum Environment of Vegans and Omnivores: https://doi.org/10.3390/nu15122755
- Leptin increases hepatic triglyceride export via a vagal mechanism in humans: https://doi.org/10.1016/j.cmet.2022.09.020
- Calorimetric approach for comparison of Angiopoietin-like protein 4 with other pancreatic lipase inhibitors: https://doi.org/10.1016/j.bbalip.2019.158553
- Apolipoprotein C-II mimetic peptide is an efficient activator of lipoprotein lipase in human plasma as studied by a calorimetric approach: https://doi.org/10.1016/j.bbrc.2019.08.130
- Lipoprotein lipase activity and interactions studied in human plasma by isothermal titration calorimetry: https://doi.org/10.1194/jlr.D071787
- Evidence for Two Distinct Binding Sites for Lipoprotein Lipase on Glycosylphosphatidylinositol-anchored High Density Lipoprotein-binding Protein 1 (GPIHBP1): https://doi.org/10.1074/jbc.M114.634626
Biomedicine
Biomedicine lab investigates Helicobacter pylori (HP) and its role in the development of liver diseases. HP is a Gram-negative bacterium living in the hostile environment of the human stomach. About 70% of the adult population in Estonia is infected with HP. The bacterium causes gastritis and peptic ulcers, and, in some cases, gastric cancer. HP can also affect other organs including the liver.
Main research topics include:
• Role of Helicobacter pylori-induced invadosomes in liver damages. We have previously shown that infection with HP induces the formation of invadosomes in hepatocytes. We are currently investigating the mechanism behind this phenomenon using in vitro approach complemented with transcriptome sequencing.
• Clinical aspects of Helicobacter pylori-induced liver diseases. We are using the livers of mice infected with HP to analyse short- and long-term effects of the infection with focus on different markers such as YAP1 and CD44.
• Alteration of gut microbiota by Helicobacter pylori leading to the progression of liver diseases. For this study, we are collecting samples from Estonian patients. Our goal is to characterize Estonian HP strains, their effect on mouth/stomach/gut microbiota and liver.
Members of the research group
Group leader: Pirjo Spuul
PhD students: Dr. Airi Rump (researcher), Irma Lill (engineer)
PhD students: Olga Smirnova, Kaisa Roots, Sadia Khalid
MSc students: Marit Kutman, Tianeke Luna Wilkie

Contact information
Pirjo Spuul, senior researcher, head of the biomedicine lab
E-mail: pirjo.spuul@taltech.ee
Address: Building of Science, Akadeemia road 15, room 140
CV: https://www.etis.ee/CV/PirjoSpuul/est?tabId=CV_ENG
DNA Replication and Genome Stability
The laboratory of DNA replication and Genome stability is working on deciphering the molecular mechanism of human replication initiation, which is both an important basic research problem and has critical implications for cancer therapy. Understanding the molecular mechanism of replication initiation in human cells will lead to developing drugs that specifically disrupt replication initiation to block the proliferation of cancer cells, or increase the number of replication forks, targeted by many chemotherapeutics, to improve the efficiency of cancer treatment.
The group currently does not accept new BSc or MSc students.
Main research topics include:
- The non-catalytic function of DNA polymerase epsilon
- TIMELESS protein in the initiation of DNA replication and cancer
- Identification of novel replication initiation factors using proximity-labelling proteomics
Members of the research group
Group leader: Tatiana Moiseeva
PhD students: Naga Raviteja Chavata
BSc students: Evelina Koop
Contact information:
Tatiana Moiseeva, senior researcher, head of the lab
E-mail: Tatiana.moiseeva@taltech.ee
Address: Natural Science Building, Akadeemia road 15, office 131A
CV: https://www.etis.ee/CV/Tatiana_Moiseeva/eng/
Glial cell biology
In addition to neurons, brains contain about the same number of specialized non-neuronal brain cells, collectively known as glia. Glial cells have several important roles in nervous system development and homeostasis, including establishing connectivity, maintenance of neurotransmission and immune functions. Together with neurons, these cells form a complex, intertwined network that is subject to constant interaction between different cell types. Our main research focus is on astrocytes, one of the principal glial cell types of the central nervous system. Among their many functions are ion and neurotransmitter homeostasis at synapses and several central metabolic roles.
The main research interests of our lab include molecular communication between neurons and astrocytes and the regulation of protein synthesis in astrocytes. For this, we make use of genetic tools for cell type-specific stimulation and gene expression analysis. Additionally, we develop methods that allow cell type-specific proteome labeling and analysis.
Members of the group:
Principal investigator: Dr. Indrek Koppel
Researchers: Dr. Florencia Cabrera Cabrera (researcher), Dr. Age Utt (researcher)
PhD students: Helena Tull
MSc and BSc students: Katariina Karm, Markus Talp, Martin Paggi, Eliisabeth Kuldmaa

Immunobiology
Sirje Rüütel Boudinot's main area of research is leukocyte activation and its regulation, which is important both in equilibrium and during the immune response. The dysregulation of immune function is a key factor in tumours, inflammatory and autoimmune diseases, and strongly influences the ability to fight pathogens.
The research group has long been involved in characterising the role of (purinergic) receptors (P2X4 and P2X7) in the initiation and progression of the immune response. Using both in vitro and in vivo models, we will perform loss-of-function and gain-of-function experiments to characterise the mechanisms mediated by these genes. We will also use comparative methods to assess the importance of these genes in the context of immune evolution. We show that the P2X7 receptor arose from the fusion of a P2X4-like receptor and the ballast domain.
In collaboration with PERH, we describe the role of P2X4 in immune cell activation and tolerance in the context of both melanoma and neuroinflammatory disorders.
We showed that glycosylation of SARS-CoV-2-NP obscures immunodominant epitopes. In many cases, this can lead to false-negative serological tests. Deglycosylation of SARS-CoV-2-NP significantly increased the number of positive tests.
Main research topics
- P2X4 expression-dependent pro- or anti-inflammatory orientation of the immune response
- Identification of the secretory form of P2X4 in melanoma and polyneoplastic neural diseases
Members
Leader of the research group: Senior Researcher Sirje Rüütel Boudinot
PhD students: Roland Martin Teras, Kadri Orro
Master students: Helinda Saapar, Anne-Lii Bôkovskaja, Adriana Kalmôkova, Igor Kuprijanov, Laura Liis Pahk, Priit Ties, Martin Frey,
Current funding
PRG1832, Development of methods for hydrogen nuclear magnetic resonance spectroscopy (H NMR).for early diagnosis of neurodegenerative diseases and to assess the impact of systemic therapy of melanoma and to study the respective molecular mechanisms.
The most important publications of recent 5 years:
Kanellopoulos J, Almeida-da-Silva CLC, Rüütel Boudinot S and Ojcius DM (2021) Structural and Functional Features of the P2X4 Receptor: An Immunological Perspective. Front. Immunol. 12:645834. doi: 10.3389/fimmu.2021.645834
Boudinot, S. (2021). Dual ELISA using SARS-CoV-2 N protein produced in E. coli and CHO cells reveals epitope masking by N-glycosylation. Biochemical and Biophysical Research Communications, 534, 457−460. DOI: 10.1016/j.bbrc.2020b.11.060
Rump, A.; Smolander, O.-P.; Rüütel Boudinot, S.; Kanellopoulos, J. M; Boudinot, P. (2020). Evolutionary origin of the P2X7 C-ter region: capture of an ancient ballast domain by a P2X4-like gene in ancient jawed vertebrates. Frontiers in Immunology, 11, 113−113. DOI: 10.3389/fimmu.2020a.00113.
Teras, J.; Kroon, H. M.; Thompson, J. F.; Teras, M.; Pata, P.; Mägi, A.; Teras, R. M.; Rüütel Boudinot, S. (2020). First Eastern European Experience of Isolated Limb Infusion for In-Transit Metastatic Melanoma Confined to the Limb: Is it still an Effective Treatment Option in the Modern Era? European Journal of Surgical Oncology. Vol 46, Feb 2020, p272-276.
Paalme, V.; Rump, A.; Mädo, K.; Teras, M.; Truumees, B.; Aitai, H.; Ratas, K.; Bourge, M.; Chiang, C.-S.; Ghalali, A.; Tordjmann, T.; Teras, J.; Boudinot, P.; Kanellopoulos, J.; Rüütel Boudinot, S. (2019). Human peripheral blood eosinophils express high level of the purinergic receptor P2X4. Frontiers in Immunology.10.3389/fimmu.2019.02074
Metalloproteomics
Palumaa Lab is focused on the studies of the biological role and the regulation of two important transition metals - copper and zinc. Copper is an essential cofactor for more than twenty enzymes crucial for cellular energy production, antioxidative defense, and oxidative metabolism. Zinc is involved in cell metabolism and regulates gene expression as it is a cofactor for more than 200 enzymes and is involved in the structuring of more than 600 transcription factors (zinc finger proteins). Dysregulation of copper and zinc homeostasis occurs in multiple diseases, including Wilson's, Menkes, and Alzheimer's disease.
Metalloproteomics group has been studying copper and zinc metabolism through structural and functional studies of key metalloproteins for a long time. In addition, in the last years, they have been using also different cellular and insect models for researching involvement of copper in Alzheimer's disease, such as cell culture and fruit flies. The expected results will substantially advance the knowledge on copper metabolism and facilitate the search for molecular tools for its regulation. This is essential for understanding the cause of Alzheimer's disease and elaboration of an effective strategy for its treatment. The research group has different methods at their disposal - LC-ICP MS for ultrasensitive detection of metals, MALDI MS, spectrofluorometer, FPLC, HPLC, and UHPLC chromatographic systems for working with proteins, etc.
Main research topics
- Regulation of copper metabolism by alpha lipoic acid.
- Identification of new copper carriers.
- Interaction of copper with tubulin
Members
Group leader: Prof. Peep Palumaa
Scientists: Prof. Vello Tõugu, Dr. Julia Smirnova, Dr. Merilin Sardis, Dr. Andra Noormägi, Anette Reinapu (MSc; engineer)
PhD students: Sigrid Kirss
MSc students: Janar Varik, Jelizaveta Kravtsova, Sofja Poddubnaja
BSc students: Hanna Parv

Rewards & collaborations
The head of the research group Prof. Peep Palumaa received the Estonian National Research Prize in Chemistry and Molecular Biology in 2011 and in 2024, the TalTech Best Researcher Prize in 2012. Our students have frequently won prizes for their thesis and publications at competitions for student research organized by Estonian Ministry of Education and Research and by research societies.
Research group collaborates actively with Stockholm University and Karolinska Institute.
Contact
Prof. Peep Palumaa
E-mail: peep.palumaa@taltech.ee
CV: Peep Palumaa | CV (etis.ee)
Microfluidics
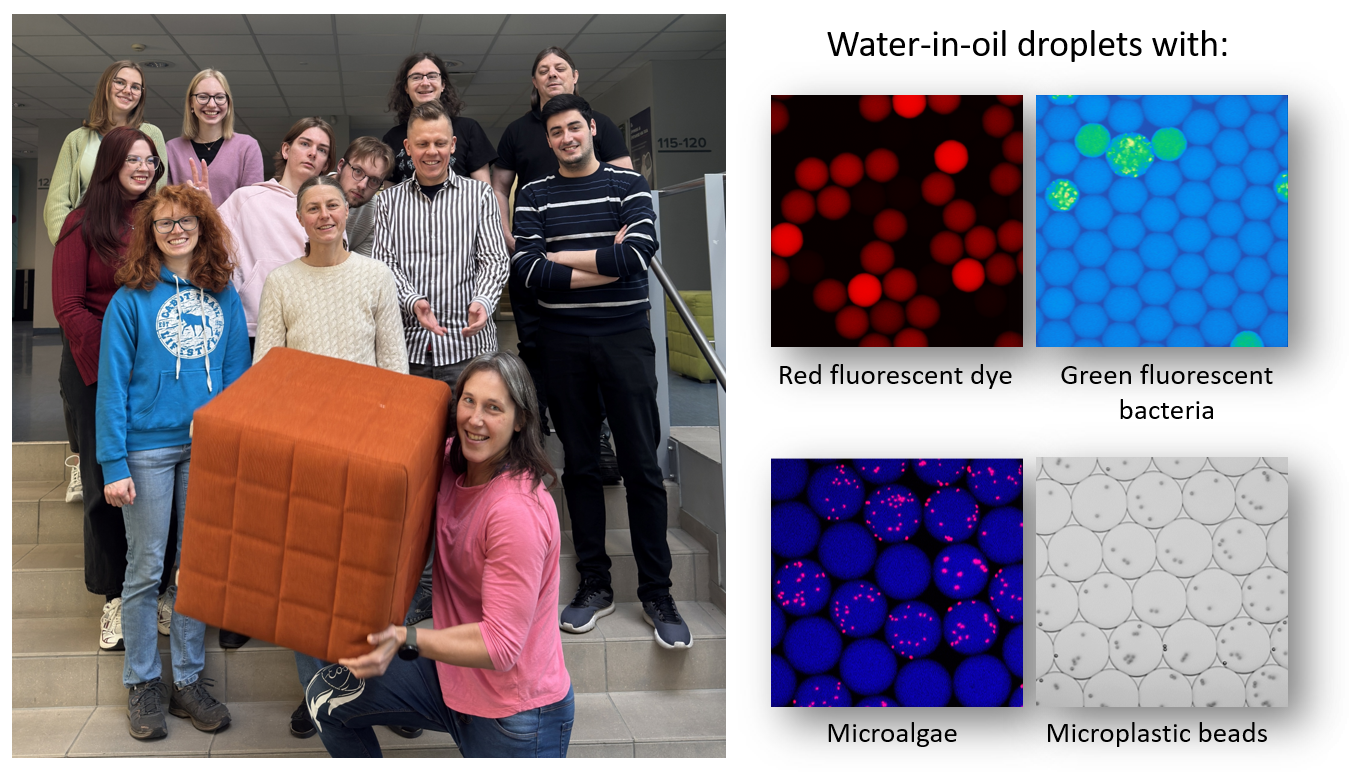
Our group is carrying out biotechnological research using different microfluidic applications. With microfluidics you can miniaturize and automate lab research in smaller experimental volumes. In our research we apply droplet microfluidics in different microbiological and biotechnological projects. With droplet microfluidics you can carry out your research in tiny water-in-oil droplets aka “test-tubes” that have diameter smaller than human hair. This is a high-throughput technology as it allows having hundreds of thousands or even more droplets in parallel in single experiment
Info for students: we are open to hosting ERASMUS+ exchanges and also help to develop novel international PhD and postdoc projects. Interested people should contact the group leader
Current research topics
- Microbiology: we apply droplet microfluidics to investigate multiple aspects of antimicrobial susceptibility and resistance. We investigate how it can be affected by different chemicals and microplastic. Additionally, we look how aggregation of bacteria and biofilm formation affect antimicrobial susceptibility patterns
- Microscopy and image analysis: we develop user-friendly workflows for the analysis of droplet microfluidic experiments. We use different microscopic approaches (Brightfield, fluorescence, confocal, etc) to visualize reactions and experiments going on inside the droplets that is followed by image analysis via different platforms (e.g. Cellprofiler)
- Lab automation: in this topic we develop devices for microfluidic and laboratory automation purposes. We do this in collaboration with Lab-on-a-Chip group led by Assistant Professor Tamas Pardy (https://sites.google.com/view/taltechloc): (micro)fluidics, hard and software for microfluidics, optics and cameras
- CellFactory generation: Together with prof. Pardy we develop droplet microfluidic tools for generation of cell factories. These are microbial or eukaryotic cells that have been modified to produce useful and interesting molecules for food- and medicine industry (enzymes, drugs, chemicals, vitamins, etc). For generation of cell factories we use droplet sorting technology (FADS)
Ongoing research projects:
- Ott Scheler: https://www.etis.ee/CV/Ott_Scheler/eng
- Simona Bartkova: https://www.etis.ee/CV/Simona_Bartkova/eng/
Members:
Principal investigator: Prof. Ott Scheler and senior researcher Dr. Simona Bartkova
Staff: Dr. Immanuel Sanka, Pille Pata
PhD Students: Veiko Rütter (ICT PhD Student co-supervised with Dr. Tamas Pardy), David Gonzalez, Daniel Kacsor
MSc students: Triini Olman, Ats Oskar Laansalu
BSc students: Inessa Bogdanova, Ksenija Nikiforova,
Visitors: Merle Nadja Zenner (Leipzig University), Robin Veere (Uppsala University)
Publications:
Molecular Neurobiology
Tõnis Timmusk has been studying the nervous system for more than 30 years, of which he has worked at Tallinn University of Technology for almost 20 years. In total, he has published more than 90 publications in high-level international scientific journals.
Today, in the laboratory of molecular neurobiology, we study the molecular basis of gene expression and signal transduction in the nervous system and its pathologies, using both mammalian nerve cells and fruit fly as a model system. We seek to understand how cells interact with each other and how this communication regulates gene expression and the connections between nerve cells – the basis of memory and learning. In addition, we are investigating the causes of one autism spectrum disorder, Pitt-Hopkins syndrome, and are looking for potential treatment possibilities.
We are innovative in our work and use modern molecular and cell biology approaches, such as CRISPR-Cas based (epi)genome modification systems, second and third generation sequencing methods, and the creation of various nervous system cells from embryonic stem cells. We also consider it important to participate in international collaborations with other research laboratories. Our goal is to develop a strong generation of neurobiologists in Estonia and we value critically thinking, motivated and enthusiastic people. The team in the neurobiology laboratory is supportive and we maintain high scientific standards.
You can find out more about the work of our laboratory at the virtual exhibition and at the virtual tour - Youtube video

Members:
Group leader: Prof. Tõnis Timmusk
Scientist: Dr. Florencia Cabrera Cabrera, Dr. Olga Jasnovidova, Dr. Laura Tamberg, Dr. Annela Avarlaid, Dr. Richard Tamme, Dr. Mari Palgi
Lab manager: Epp Väli
PhD students: Anastassia Šubina, Carl Sander Kiir
Students: Liis Kuusemets, Artur Morgunov, Markus Jonathan Mõis
Neuroepigenetics
The laboratory of neuroepigenetics studies how epigenetic mechanisms control neuronal development and function throughout life. Neurons are born and mature early in development and are typically not replaced during the organism's lifetime. Therefore, once neurons have matured and become fully functional, they must remain so for a long time - in humans, for up to a hundred years or more. This means that the transcriptional programs that become established during neuronal maturation must remain stable - yet responsive to external stimuli - over the entire lifespan of an organism. We are interested in how the epigenetic landscape is set up and controls transcriptional programs in during neuronal maturation and how epigenetic and transcriptional stability are maintained in neurons throughout life.
Key projects
- Histone bivalency in neuronal development and function
- The role of H3K27me3 in adult and ageing neurons
Members
Principal investigator: Dr. Kärt Mätlik
PhD students: Irma Laas
BSc students: Liisi Mets, Sarah-Elisabeth Soitu

Contact
Dr. Kärt Mätlik
E-mail: kart.matlik@taltech.ee
Protein design
Proteins are biopolymers with a myriad of properties and functions: for instance, keratin is the key structural material in our hair and nails, hemoglobin transports oxygen in our blood stream, and digestive enzymes break down food into absorbable components. These diverse functions are directly related to protein structure. The goal of the Protein Design Lab is to understand these structure-function relationships in order to develop new proteins with enhanced properties for the benefit of humanity.
Our main research areas:
- Development of novel protein-based food additives (e.g., colorants and sweeteners)
- Rational and AI-driven protein design
- Structural analysis, including X-ray crystallography
Members
Principal investigator: Dr. Priit Eek
Scientists: Dr. Kaia Kukk, Dr. Hegne Pupart
PhD students: Maarja Lipp, Azra Melkić, Balamanikandan Saptharishi
Master student: Liis Hirvesoo
Bachelors students: Reio Pukonen, Riina Tannil, Taissia Zayvaya

Contact
Dr. Priit Eek
E-mail: priit.eek@taltech.ee
CV: https://www.etis.ee/CV/Priit_Eek/eng/
Reproductive Biology
Infertility is a global problem affecting approximately 15% of all couples in a fertile age. While there are many causes for both male and female infertility, possible treatments are fortunately similarly numerous. One common method enabling otherwise infertile couples to receive genetically their own offspring is in vitro fertilization (IVF). Sadly, the efficiency of IVF is low – on average only one in three procedures culminates in the birth of a child.
The Research Group for Reproductive Biology studies the molecular mechanisms behind fertility and infertility. We are focused foremost on female infertility and on such ovarian processes which assure the maturation of a healthy egg cell. This maturation is affected by hormones from the pituitary gland, steroid hormones produced by the ovaries and several different signaling molecules that move between the egg cell and the granulosa cells surrounding it. By identifying key signaling pathways for egg cell maturation in granulosa cells we can use these cells for diagnostics and thus greatly improve the effectiveness of the IVF procedure.
The ovarian follicle is the main research object in our group. As the oocyte is used for fertilization and embryo development during IVF procedures, invasive methods to study its ingredients are not preferable. However, the environment of oocyte maturation can be evaluated by investigating its surrounding granulosa cells and the follicular fluid that contain a lot of genetic and biochemical information.
In our work we use both classical laboratory methods as well as methods for high throughput analysis (such as analyzing gene expression by deep sequencing, microchip or mass spectrometry methods). In addition, we perform bioinformatical data analysis, biostatistics and modelling signaling pathways.
We are in close cooperation with the Competence Centre on Health Technologies (Tervisetehnoloogia Arenduskeskus AS) and with all fertility clinics in Estonia.
Group members:
Principal investigator: Associate professor Agne Velthut-Meikas
Scientist: Dr. Airi Rump, researcher
PhD students: Inge Varik, Laura Luhari, Mayara Cristina Batista Da Silva, Katariina Johanna Saretok
MSc students: Thor Tristan Karafin
BSc students: Johanna Rannaveer, Ksenia Ponomarjova

Smart Analytics
We join the specialists from various departments, universities, and private sector, aiming to ensure groundbreaking research can be turned into successful business opportunities. Over the last 15 years we have successfully developed various analyzers (TRL6-7) for different partners (e.g. Estonian Police and Border Guard). Our core technologies are Capillary Electrophoresis, Fluorescence, Conductivity, Gas Chromatography, Microfluidics, and other instrumental and analytical techniques. We collaborate with various research groups worldwide, providing our competence for successful participation in Horizon Europe and other various open calls.
Visit Drug Hunter Analyzer’ website for more information about drug analysis in oral fluid www.drughunter.eu
People
Group leader - Dr. Jekaterina Mazina-Šinkar (jekaterina.mazina@taltech.ee),
Dr. Jelena Gorbatšova,
Dr. Evelin Halling,
Professor emeritus Mihkel Kaljurand,
Dr. Merike Vaher,
Dr. Martin Ruzicka,
Vyacheslav Bolkvadze,
Jana Budkovskaja